NucleoSpin Food, Mini kit for DNA from food
*taxes and shipping not included
NucleoSpin Food is a spin column based kit for manual use. It is intended for the isolation of DNA from processed and unprocessed food and feed. Given the wide range of possible samples, the kit is robust against high acidity or alkalinity, contaminant-rich and hard-to-lyse samples.
| Application | Isolation of DNA |
| Selling unit | 10 Prep(s), 50 Prep(s), 250 Prep(s) |
| Target | DNA |
| CE certified | No, research use only |
| Technology | Silica membrane technology |
| Brand | NucleoSpin |
| Format | Mini prep |
| Handling | Centrifugation |
| Lysate clarification | Centrifugation |
| Automated use | No |
| Sample material | Feed, Food |
| Sample amount | < 200 mg |
| Fragment size | 300 bp–approx. 50 kbp |
| Typical yield | 0.1–10 µg (200 mg food) |
| Theoretical binding capacity | 30 µg |
| Typical purity A260/A280 | 1.6–1.9 |
| Elution volume | 100 µL |
| Preparation time | 30 min/6 preps |
| Typical downstream application | enzymatic reactions, Real-time PCR, Southern blotting |
| Storage temperature | 15–25 °C / 59–77 °F |
| Shelf life (from production) | 27 Month(s) |
| Hazardous material | Yes |
NucleoSpin Food
- Complete removal of PCR inhibitors – get high quality DNA
- Even low amounts of partially degraded DNA can be purified from complex matrices
- Fast and easy procedure
- DNA from various sample materials – highest flexibility
NucleoSpin Food - Procedure
Identification of GMO-DNA or animal components in food and feed has become an important application of public interest. In the novel food regulation, labelling has been mandatory for GMO containing food since they can be distinguished from conventional products by analytical methods, for example real-time PCR. Food samples are very heterogeneous and contain many different compounds like fat, cocoa or polysaccharides which can lead to suboptimal extraction or subsequent processing of DNA. NucleoSpin Food guarantees good recovery rates even for small genomic DNA fragments (< 1 kb) out of processed, complex food matrices (e.g., ketchup or spices), which generally have very low DNA contents as well as poor quality, degraded DNA.
After the food samples have been homogenized, the DNA can be extracted with lysis buffers containing chaotropic salts, denaturing agents and detergents. The standard isolation ensures lysis using Buffer CF, which was especially developed in cooperation with GEN-IAL, Troisdorf, Germany (patent pending). Lysis mixtures should be cleared by centrifugation or filtration in order to remove contaminants and residual cellular debris. The clear supernatant is then mixed with binding buffer and ethanol to create conditions for optimal binding to the NucleoSpin silica membrane, which was selected for this purpose due to its unique DNA-binding properties.
After washing with two different buffers for efficient removal of potential PCR inhibitors, DNA can be eluted in low salt buffer or water, and is ready-to-use in subsequent reactions.
Reliable extraction of DNA from various seed matrices
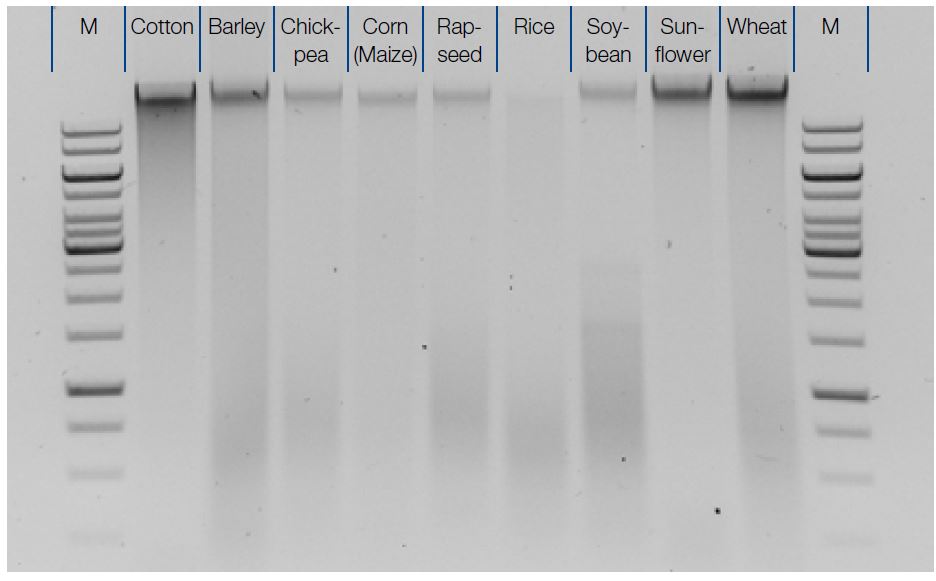
After spectrophotometric analysis, DNA purified from several seed samples was further examined by agarose gel electrophoresis. For each sample 2 µL of eluate were loaded onto a 1 % agarose gel. The marker (M) was 1 kB ladder (Fermentas). While the amounts of extracted DNA varied between different sample types, in each case detectable amounts of DNA could be reliably extracted from 200 mg of sample.
Amounts and quality of the extracted DNA were sufficient for detection by qPCR
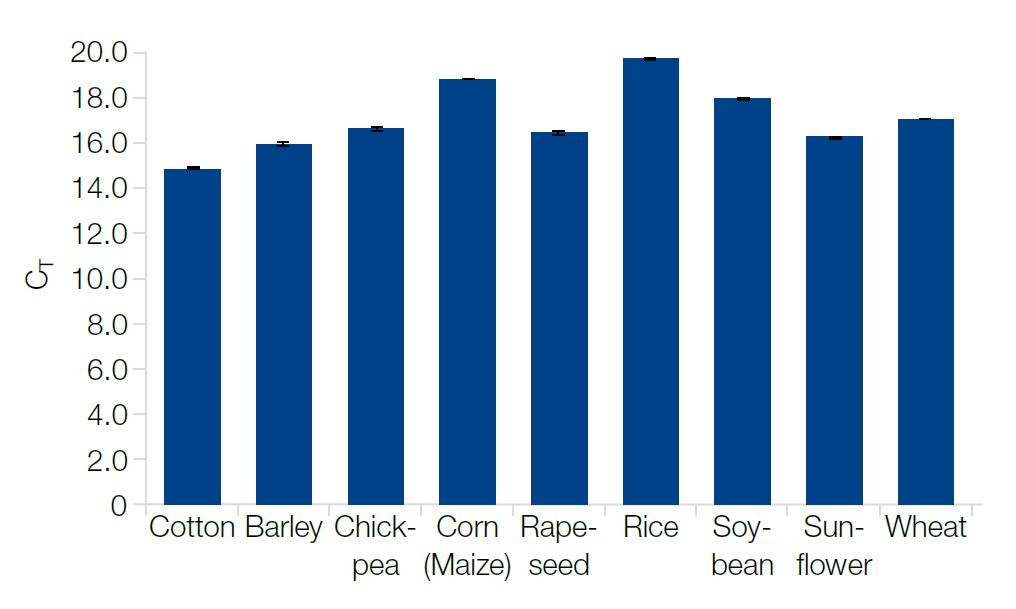
The extracted samples were analyzed by qPCR with the Eu +/- marker. The purity and concentration of DNA were sufficient for reliable detection of the genetic marker and thus suitable for genotyping.
GMO, food and feed analysis with NucleoSpin Food
NucleoSpin nucleic acid isolation technology from MACHEREY-NAGEL and GMO experience from GEN-IAL were combined to provide an optimal lysis and purification system for nearly all types of food samples. Resulting eluates are ready-to-use for all types of subsequent detection methods, especially real-time and basic PCR technology.